L519: Lab Session 10 (11/11/05)
- TRANSFAC® is the database on eukaryotic transcription factors, their genomic binding sites and DNA-binding profiles. It covers the whole range from yeast to human. Public version is freely available.
- TRANSFAC® Tables and their
Relations.
SITE
7915 entries
GENE
2397 entries
(1504 entries with SITE links) FACTOR
6133 entries
CELL
1307 entries
CLASS
50 entries
MATRIX
398 entries
- The contents of each table will be
detailed below.
SITE : information on (regulatory) transcription factor binding sites within eukaryotic genes.
GENE : short explanation of the gene where a site (or group of sites) belongs to.
FACTOR : the proteins binding to these sites.
CELL : brief information about the cellular source of proteins that have been shown to interact with the sites.
CLASS : some background information about the transcription factor classes
MATRIX : table gives nucleotide distribution matrices for the binding sites of transcription factors.
- Public version: http://www.gene-regulation.com/pub/databases.html#transfac
- IU Professional version: http://bioportal.cgb.indiana.edu/ (Account is required)
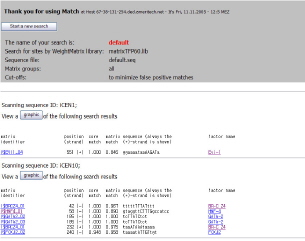
- MatchTM is designed for searching potential binding sites for transcription factors (TF binding sites) nucleotide sequences. MatchTM uses a library of mononucleotide weight matrices from TRANSFAC® 6.0. (Matrix)
- P-MatchTM is new tool for identifying transcription factor binding sites (TF binding sites)
in DNA sequences. It combines pattern matching and weight matrix approaches thus
providing higher accuracy of recognition than each of the methods alone. P-Match
uses a library of mononucleotide weight matrices from TRANSFAC® 6.0 along with
the site alignments associated with these matrices.
- Sample Sequence : 5 Yeast Promoters
Match Result : SampleMatch.html
P-Match Result : SamplePMatch.html
- Try the following set of sequences : SampleSeqSet
- Do you remember what the EST_GENOME is?
- This was briefly introduced during the Lab 7 with GenScan and TwinScan.
- http://www.well.ox.ac.uk/~rmott/ESTGENOME/est_genome.shtml
- Source code (needs to be modified to be compiled) and executable version are available on the biokdd.
/tmp/L519FALL2005/EST_GENOME
You better copy it to your own directory if this program is needed for your project.
- Commandline parameters

You might have to use full path if you don't have it specified in the path.
> /tmp/L519FALL2005/EST_GENOME/bin/est_genome - Sequence format : FASTA
- Number of sequences in the FASTA file
- genome : single sequence
- est : multiple sequences
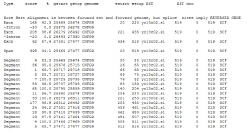
- Sample usage
> est_genome -genome CNFG9.seq -est yo13c02.s1.seq
Result report
- Full alignment : with -align option

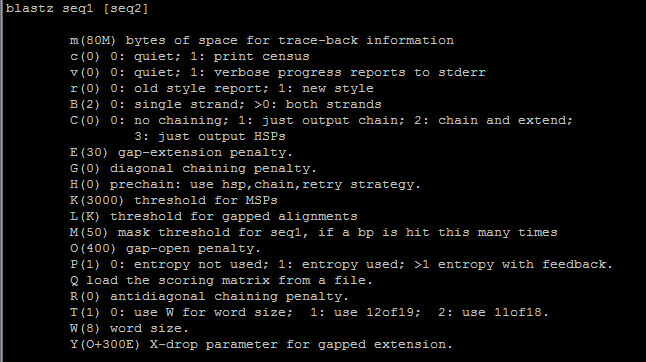
- BLASTZ is a pairwise sequence alignment program for the whole-genome alignments.
- Blastz output can be viewed with the LAJ interactive alignment viewer, converted to traditional text alignments using LAT, or you can write a program to parse it.
- BLASTZ is found at /usr/local/biokdd/bin/blastz.
- options are as follows. Default values are given in parentheses
- The command line would be like
> blastz <seq1> <seq2> B=0 K=2200 > blastz.out - Genome sequences are available on the biokdd
/data/biokdd/genbank/GENOMES/ - Sample genome sequences are in the tmp directory (/tmp/L519FALL2005/Lab10/)
- CNFG9.seq : Portion of Human Chromosome 16. (We used for EST_GENOME)
- Helicobacter_pylori_26695.fna : Helicobacter pylori strain 26695 (causing ulcer)
- Helicobacter_pylori_J99.fna : Helicobacter pylori strain J99 (causing ulcer)
- Mycoplasma_genitalium.fna : parasite in primate genital and respiratory tracts. smallest genome (other than virus) 580 kbp.
- Mycoplasma_pneumoniae.fna : Causing pneumoniae in human
- Mycoplasma_pulmonis.fna : Infecting rat and mouse
- Try some pairwise genome alignments
- > blastz Mycoplasma_pneumoniae.fna Mycoplasma_pulmonis.fna > pneumoniae_pulmonis.blastz.out (1 minute on biokdd)
- Here some results
- genitalium_pnumoniae.blastz.out 1strand_comp
- genitalium_pulmonis.blastz.out
- pnumoniae_pulmonis.blastz.out
- Helicobacter_pylori_26695_J99.blastz.out
- genitalium.SELF.blastz.out
- Alignment result viewers and other programs are available at Miller's lab at Penn State University.
- Laj : Laj interactive viewer for pairwise alignments
- Scott Schwartz, W. James Kent, Arian Smit, Zheng Zhang, Robert Baertsch, Ross C. Hardison, David Haussler, and Webb Miller Human-Mouse Alignments with BLASTZ Genome Res., Jan 2003; 13: 103 - 107.
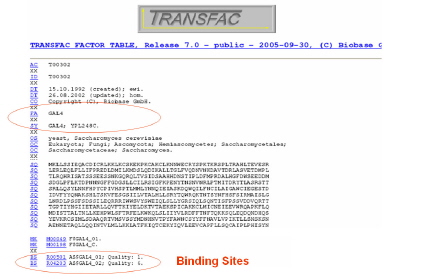
- Search for the matrix of GAL4 transcription factor in TRANSFAC
- The following sequences are known to have binding sites for a specific TF.
- Use Match and P-Match to see what the possible TF is.
- Did you find any consistent occurrence of specific TF?
- Let's use the motif finding programs we saw last week (MEME, Gibbs sampler)
- Run MEME and Gibbs on the sequence set.
- If not, lower the threhshold
- Compare the candidate motifs with your finding from TRANSFAC.
- * Don't forget to give options to use DNA sequence and reverse strands
- If you find a motif that is consistent among the results, create an image file using Logos
Last Modified : November 11, 2005